In this 2-hour hands-on workshop, we will provide an overview of selected methods for graphical model estimation and network modeling, with application to metabolomics. Metabolomic data can be effectively represented as networks, in which nodes represent individual metabolites and edges between pairs of nodes represent dependencies between metabolite pairs. Within this framework, we will describe methods for graphical model estimation such as the gLasso (Hastie, T. and Tibshirani, R., 2008) that are useful for describing the conditional relationships between metabolite pairs in a p>n setting. We will also highlight methods for multiple group graphical model estimation (Danaher, P. et al., 2014) and differential network analysis, such as DINGO (Ha, M. J. et al., 2014). Examples will be provided highlighting application of these techniques in clinical research.
Workshop Objectives
Visualize network models using R
Apply R programming to analyze metabolomics network data
Explain the use of network models in metabolomics studies
Workshop Outcomes
Perform differential network analyses using R
Create R objects for network modeling using the igraph R package
The workshop will include hands-on R exercises in addition to didactic training. In preparation for the workshop, please download R and the required R packages onto your laptop as described here:
Download InstructionsNote on Installation:
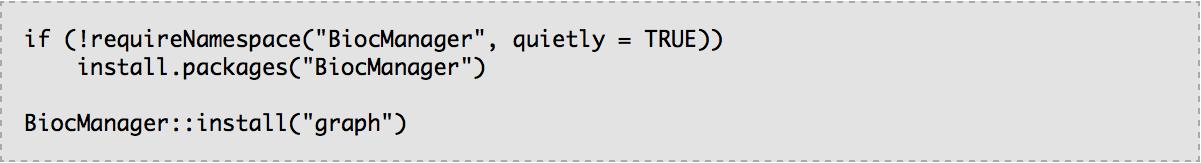
R package graph is available on Bioconductor. In R console, enter the following:
After installing R and the required packages, please download the following .zip archive and store all files in a directory named ‘Metabolomics Workshop 2019’ on your desktop:
Download ZipIndividual files included in the .zip file are also available for download here:
Download Dataset Download rda Download R Code
(Part 1) Download R Code
(Part 2) Download Slides
(Part 1) Download Slides
(Part 2)